
Predicting the risk of drug-induced liver injury using the polygenic risk score (PRS)
Genomic stratification of drug-induced liver injury (DILI)
T-CiRA’s Science Series
In the new drug development process, the drug’s efficacy and safety, as well as the appropriate dose, are investigated first in preclinical studies using cells or other materials, and then in clinical trials with healthy individuals and patients. There are many cases where safety problems come to light in the clinical trial stage, and the drug’s development is then terminated. One of the leading causes of this is drug-induced liver injury. Therefore, the precise prediction of liver injury would be of enormous benefit to drug development and medical care. T-CiRA has worked on a method for determining the risk of liver injury from genome information and expressing it as a numerical score. We have also carried out experiments using liver organoids created from iPS cells, as well as hepatocytes, to confirm the usefulness of the risk score in predicting disease onset.
・Publication
Polygenic architecture informs potential vulnerability to drug-induced liver injury
Masaru Koido, Eri Kawakami, Junko Fukumura, Yui Noguchi, Momoko Ohori, Yasunori Nio, Paola Nicoletti, Guruprasad P. Aithal, Ann K. Daly, Paul B. Watkins, Hisashi Anayama, Yvonne Dragan, Tadahiro Shinozawa & Takanori Takebe
Nat Medicine (2020). https://doi.org/10.1038/s41591-020-1023-0
Susceptibility to drug-induced liver injury is associated with numerous gene polymorphisms
Drug-induced liver injury is one of the leading causes for terminatng drug development. It may take pharmaceutical companies as long as more than 10 years to develop a new drug, with massive investments of personnel and money. Thus, drug development termination because of the occurrence of drug-induced liver injury is a huge blow to the company, as well as a great loss for society. Failure of drug development at the clinical trial stage is particularly costly, and researchers have therefore been keen to avoid this by finding ways of predicting the risk of liver injury in the preclinical stage.
The relationship between the genome and susceptibility to drug-induced liver injury is being analyzed by some international organizations. They have so far collected genome information from people who suffered liver injury from over 150 drugs. From this data analysis, genetic polymorphisms (regions of a gene that have different base sequences in different people) that are specific to people who have suffered drug-induced liver injury have been identified. We have shown that the polygenic score, which sums up the effects of numerous SNPs, gives information about potential DILI susceptibility in humans.
We used the data from these international organizations to devise a polygenic risk score (PRS), and we examined whether this score could actually be used to predict liver injury. The PRS was calculated to determine the association between the risk of disease and multiple gene polymorphisms, rather than a single polymorphism. By examining the relationship between disease status and genetic polymorphisms in a large number of patients, the “weight” of the effect of individual genetic polymorphisms on disease states can be determined. This weight is then used to calculate the PRS from the genome information of the people or cells under study. In this study, we used data from more than 800 patients to determine the weights of more than 20,000 genetic polymorphisms in the genome, which we then used to calculate the PRS.
Prediction of liver injury using liver organoids from iPS cells
We first investigated whether the PRS could be used to predict liver injury in preclinical studies. In preclinical studies, the safety of drug candidates is examined by exposing cells and other materials to them. We carried out experiments in which we used liver organoids created from human iPS cells with known genome information (Fig. 1), as well as human primary hepatocytes derived from liver tissue, also with known genome information.
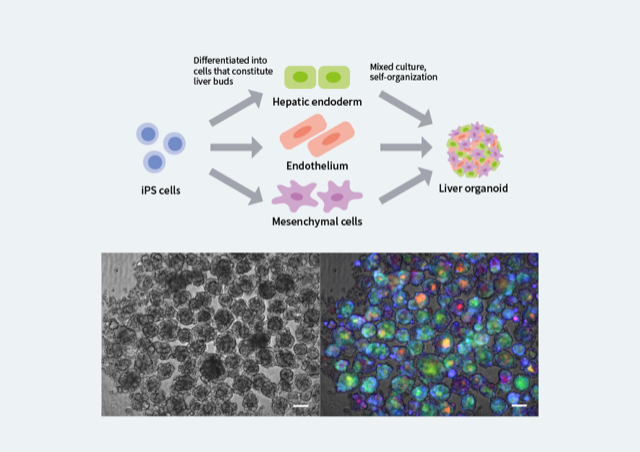
Figure 1. Generation of liver organoids from iPS cells
Liver organoids can be created by culturing together the three types of cells that are the basis of the liver. The organoids are 3-dimensional, and they have multiple structures such as blood vessels. Because multi-donor iPS cells with whole genome sequence data are used, the genome information of the liver organoid is already known. The left image shows a grouping of liver organoids observed under a microscope. The right image shows the fluorescent colors indicating the survival state of the cells (red = cell death, blue = oxidative stress, green = bile acid accumulation). The scale bar is 100 µm.
In the experiment, we administered 12 drugs known to cause liver injury to the liver organoids and to the hepatocytes, and we assessed cell viability by imaging and measuring ATP production (Fig. 2). At the same time, we also calculated the PRS from genome information. We found that the higher the PRS, the greater the decrease in the cell survival rate, and this result indicates that the PRS can be used in the preclinical stage to predict the likelihood of drug-induced liver injury. This method can be expected to lead to a user-friendly system for drug safety evaluation.
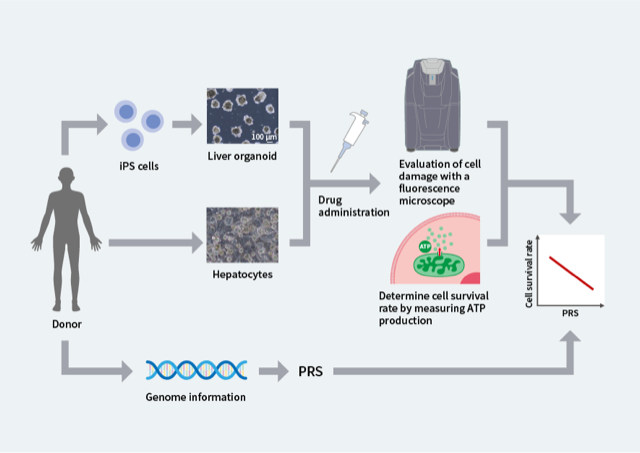
Figure 2. Confirmation of the relationship between PRS and liver injury through experiments using liver organoids and hepatocytes
Drugs were administered to liver organoids derived from iPS cells and to primary human hepatocytes, and the survival rate was observed. At the same time, the PRS was calculated from the genome information of the organoids and the hepatocytes. With all of the drugs that were investigated, a higher PRS was associated with a lower cell survival rate.
Possible clinical application of the PRS to predict disease occurrence
Next, we investigated whether it is possible to predict the occurrence of drug-induced liver injury from the PRS in clinical cases, rather than just in experiments. The individual PRSs of people who suffered liver injury during clinical trials of three drugs were calculated using their genome information. These scores were compared to those of people who suffered no effects from the same drugs, and the results showed that the PRS was clearly higher in people who had suffered liver injury.
“If we know that people with a particular genome are more prone to liver injury, it will be easier for the doctors treating them to select the right drugs,” explains Eri Kawakami, the Principal Scientist leading this research. “If we can predict whether a drug we intend to use is likely to have side effects, we might then be able to use the PRS as a diagnostic aid for side effects.” Another possible use of the PRS would be to exclude people with a high PRS from participating in clinical trials of new drugs, thus avoiding the risk of drug development ending in failure.
T-CiRA also examined which genes are associated with susceptibility to liver injury, and we found that there appears to be a particular association with genes that work during times of oxidative stress. This means that people with a high PRS are likely to be more susceptible to oxidative stress, and combining drug administration with antioxidants may prevent side effects.
Kawakami has high hopes for future development. “It looks like the PRS will be useful for predicting and avoiding not just liver injury, but also a whole range of other side effects from drugs,” she says. “We may also be able to use it to evaluate drug efficacy if we know that a drug will be effective in one particular type of person and will be ineffective in a different type of person.”